The Bio-Science AI Hackathon
Introduction
Recently, I had the chance to join the QBI bio-hackathon at UCSF. This year’s focus was on using machine learning and artificial intelligence to advance medicine. There were many incredible projects, like breast cancer detection using imaging and protein visualization. I was excited about contributing my computing skills to medical problems. Technology advancements that improve medicine can significantly enhance people’s lives.

Initially, I didn’t have a team. Fortunately, we were given the opportunity to pitch ideas and join existing teams. One pitch that caught my attention was from a Ph.D. researcher at UCSF, who was grappling with a challenging issue related to scientific data.
Advancements in Artificial Intelligence and Machine Learning in medicine depend on high-quality data for successful results. For example, Alpha Fold, arguably the most successful AI tool in biology to date, relied on the Protein Data Bank, a manually curated database of biomolecular structures for training. This data bank was assembled by researchers throughout the field collaborating and contributing in this centralized repository. Thus, for further predictive analysis capabilites, we need more data.
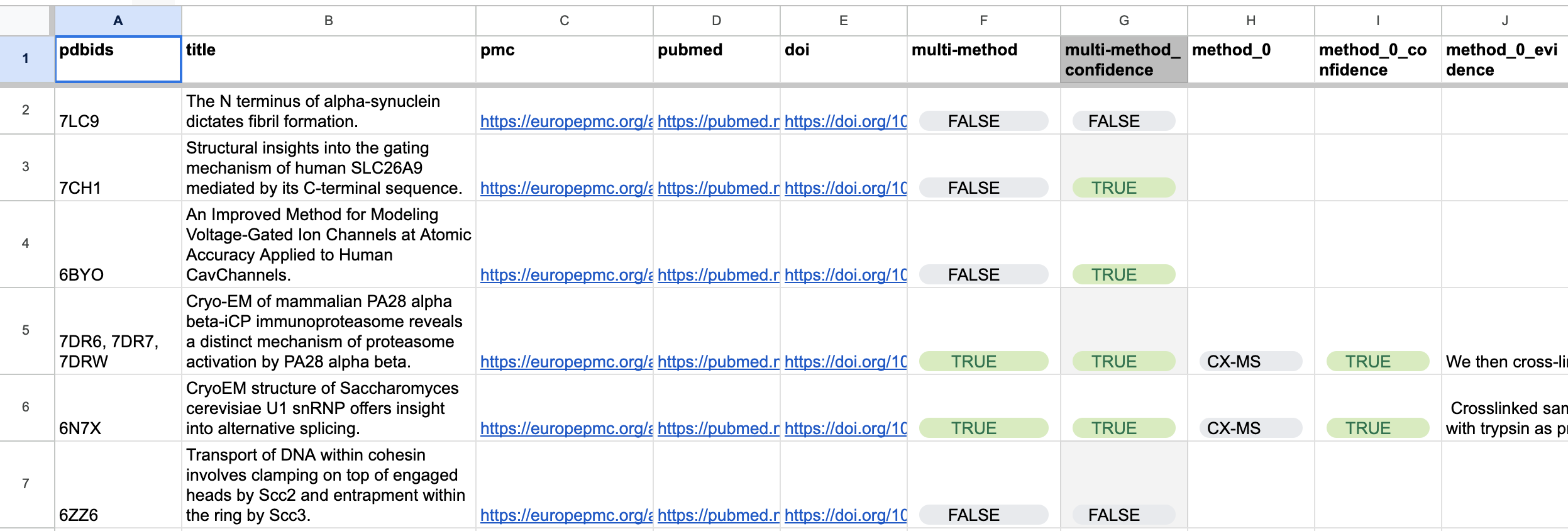
However, getting the correct data and metadata is difficult, as researchers do not like spending time on data formatting and deposition. Oftentimes, they may not be aware or simply forget to input the data into the data bank whenever they have discovered a new structure. Thus, to solve this issue, we want to scan through recently published papers that report structures of biomolecules so we can reach out to authors and bring their data into PDB.
The researcher leading this hackathon project, over the previous summer, had interns manually go through papers and fill out a spreadsheet with information about the article. Such information includes which methods were used to image the protein or if multiple methods were used. The team could then reach out to the researchers and encourage/collaborate with them to input their data into the protein data bank. The given task, for our hackathon, was to try and automate this pipeline. With recent advancements using LLMs, we figured it’d be possible to do the entire pipeline.
Project Description
Here were the specific goals we were tasked with:
- A Python script that accepts a paper (plain text or pdf) and performs annotation with a set of predefined questions.
- A Python script, that pulls updates from a set of bio/medRxiv subtopics, calls script 1, puts results into a GoogleSheet and triggers email notification.
- Purely open source/free software implementation using open/free LLMs or other approaches.
Since the relevant papers are available through bioRxiv and medRxiv, we used those as the data source. They conveniently have an API to access journal data as well.
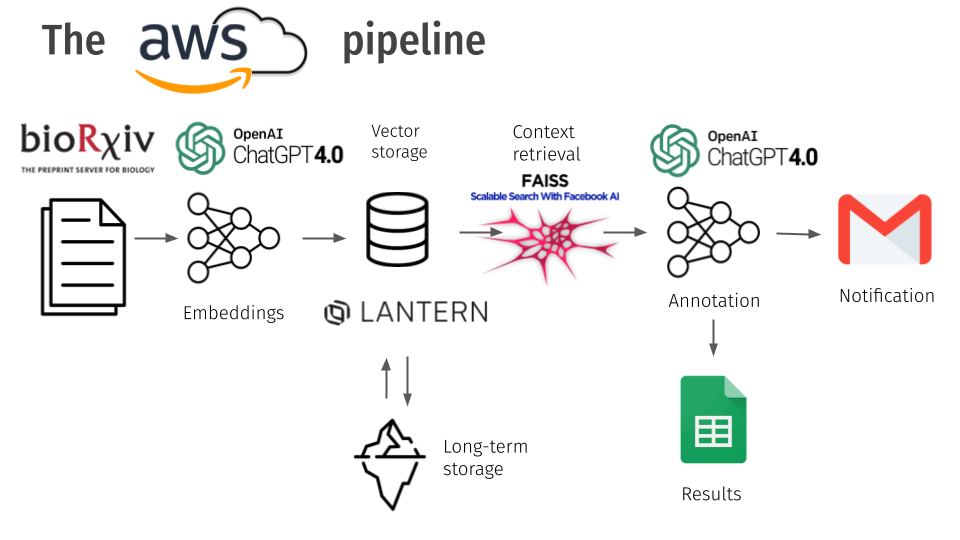
To get the prototype working, we decided to use the GPT 4.0 API. While this does break the 3rd goal, the LLM is easily replaceable by an open source alternative. The basic pipeline we created is as follows. We get the papers from the bioRxiv API and extract the metadata from the content. We turn the content into vector embeddings using OpenAI’s embeddings and store the embeddings in the Lantern Vector+postgres database, who was sponsoring the event. This helped us store the data about the papers for the long term and ask additional questions if necessary. We then used FAISS context retrieval to retrieve the context based on a given input using cosine similarity. We use GPT 4.0 to annotate the retrieved data and use Google Sheets and Mail APIs to save the data and notify stakeholders. We use AWS to host the architecture through an EC2 instance. We used langchain to help with LLM utilities.
Implementation
I will now go into depth for each part of the architecture with code. Starting with the bioRxiv scraper, in scraper.py: We utilized a python library that provided the bioRxiv API functionality for us called paperscraper.
This allowed us to use the function bioXriv to download pdf versions of papers published between a certain date.
"""
Scrapes papers from bioRxiv between the specified dates and saves the metadata in a JSON file.
:param start: Start date for the scraping (format: "YYYY-MM-DD").
:param end: End date for the scraping (format: "YYYY-MM-DD").
:param out_file: Output file to save the metadata in JSON Lines format.
:return: None
"""
def scrapeBiorxiv(start, end, out_file):
filepath = out_file
biorxiv(begin_date=start, end_date=end, save_path=out_file)
retreiveTextFromPdf(filepath)
After downloading all the papers, we scrape them using the retreiveTextFromPdf function. For each unique article (identified by DOI stored in the SQL database), we extract the text from the pdf, and get vector embeddings using the get_embeddings() function.
"""
Retrieves text from PDF files, extracts embeddings, and stores information in a custom database.
:param inp_file: Path to the input JSON file containing paper metadata.
:return: None
"""
def retreiveTextFromPdf(inp_file):
json = pd.read_json(path_or_buf=inp_file, lines=True)
lantern = Lantern()
for n, doi in enumerate(json['doi']):
paper_data = {'doi': doi}
doi = doi.replace("/", "-")
if lantern.publicationExists(doi):
continue
pdf_dir = './papers/'
if not os.path.exists(pdf_dir):
os.mkdir(pdf_dir)
pdfsavefile = './papers/' + doi + '.pdf'
save_pdf(paper_data, filepath=pdfsavefile)
# creating a pdf reader object
reader = PyPDF2.PdfReader(pdfsavefile)
save_txt_path = 'scrapped_txts/'
if not os.path.exists(save_txt_path):
os.mkdir(save_txt_path)
extract_text = ''
for page in reader.pages:
extract_text += page.extract_text()
txt_file = str('{}.txt'.format(doi))
with open(save_txt_path + txt_file, 'w') as file:
file.write(extract_text)
txt_embs, emb = get_embeddings(save_txt_path + txt_file)
...
Get_embeddings will split the input text into chunks and create a vector embedding using OpenAI Embeddings for each chunk.
"""
Retrieves text embeddings from a given text file using OpenAI's language model.
:param fname: Path to the input text file.
:return: A tuple containing text embeddings and the OpenAIEmbeddings instance.
"""
def get_embeddings(fname):
loader = TextLoader(fname)
documents = loader.load()
text_splitter = CharacterTextSplitter(
separator=".", chunk_size=1000, chunk_overlap=0)
docs = text_splitter.split_documents(documents)
emb = OpenAIEmbeddings()
input_texts = [d.page_content for d in docs]
input_embeddings = emb.embed_documents(input_texts)
text_embeddings = list(zip(input_texts, input_embeddings))
return text_embeddings, emb
To get an understanding of the next part, we have to understand the database structure. We decided to have to main tables with the following structures:
- The Publication Table - Holds publication metadata
- DOI (Digital Object Identifier) - Universal Paper Identifier
- The title
- pmc link
- pubmed link
- The Fragments Table
- DOI (Foreign Key used to identify which paper fragment is from)
- Header
- Content
- Vector
# Class to represent a publication with attributes id, title, pmc, pubmed, and doi
class Publication:
id = ""
title = ""
pmc = ""
pubmed = ""
doi = ""
def __init__(self, id, title, pmc, pubmed, doi):
self.id = id # (DOI) Unique identifier for the publication
self.title = title # Title of the publication
self.pmc = pmc # PubMed Central (PMC) Link
self.pubmed = pubmed # PubMed Link
self.doi = doi # Digital Object Identifier (DOI) Link for the publication
# Class to represent a fragment of a publication with attributes id, header, content, and vector
class Fragment:
# Class variables to store default values for attributes
id = ""
header = ""
content = ""
vector = ""
def __init__(self, id, header, content, vector):
# Constructor to initialize the attributes of the Fragment object
# Set the attributes of the object with the values provided during instantiation
self.id = id # (DOI) Unique identifier for the fragment
self.header = header # Header or title of the fragment
self.content = content # Content or text of the fragment
self.vector = vector # Vector representation of the fragment
After we have all the embeddings, we create a list of Fragments and a Publication which we then store in the database using insertEmbeddings and insertPublication. These are simple wrappers on insert commands into the database.
def retreiveTextFromPdf(inp_file):
...
fragments = []
for txt, embs in txt_embs:
fragment = Fragment(doi, 'methods', txt, embs)
fragments.append(fragment)
publication = Publication(doi, title, pmc, pubmed, doi)
lantern.insertEmbeddings(fragments)
lantern.insertPublication(publication)
os.remove(pdfsavefile)
This code has the ability to be run automatically once a day through a script or with a fixed timespan if run manually.
if __name__ == "__main__":
# Adding command line arguments for start_date and end_date with default values as the current date
parser = argparse.ArgumentParser(description="Scrape and process scientific papers from bioRxiv.")
parser.add_argument("--start-date", default=str(datetime.date.today()), help="Start date for the scraping (format: 'YYYY-MM-DD').")
parser.add_argument("--end-date", default=str(datetime.date.today()), help="End date for the scraping (format: 'YYYY-MM-DD').")
parser.add_argument("--outfile", default="bio.jsonl", help="Output file to save the metadata in JSON Lines format.")
args = parser.parse_args()
# Calling the scrapeBiorxiv function with command line arguments
scrapeBiorxiv(args.start_date, args.end_date, args.out_file)
The next part of the program is the document analyzer, which automates the evaluation of publications. This component of the system is designed to determine whether a paper mentions specific structural biology methods and subsequently updates the tracking systems with the results. The document analyzer is encapsulated in the DocumentAnalyzer class. It integrates with several services, including the Lantern vector database, Google Sheets for result tracking, and our custom LlmHandler class for handling interactions with the large language model (LLM).
For every unread publication in the database, the text embeddings are retrieved to check if the paper is about cryo-EM, analyze the publication if it is, then update the spreadsheet with the results, and send an email notification if configured to do so.
"""
pulls all new files from Lantern database, evaluates them, and publishes results to google sheets
"""
def analyze_all_unread(self):
publications = self.lantern.getUnreadPublications()
self.process_publications(publications)
def process_publications(self, publications: [Publication]):
"""takes a list of publications, applies retrievalQA and processes responses
Args:
publications ([]): list of publications
"""
rows = []
hits = 0
for pub in publications:
text_embeddings = self.lantern.get_embeddings_for_pub(pub.id)
classification, response = 0, ''
if self.paper_about_cryoem(text_embeddings):
classification, response = self.analyze_publication(text_embeddings)
hits += classification
else:
# print('paper not about cryo-em')
pass
# add date if it's added
rows.append([pub.doi, pub.title, "", str(date.today()), "", int(classification), response, ""])
self.update_spreadsheet(rows, hits)
We know its unread since we used a separate table that only contains the id of publications that were inserted after the last read.
"""
Retrieves unread publications from the 'publications' table.
Parameters:
- delete_unread_entries: bool, decides if entries are deleted from the "unread" table
Returns:
- List[Publication], a list of Publication objects representing the unread publications.
Notes:
- Performs a left join between 'publications' and 'unread' tables to retrieve unread publications.
- Clears the 'unread' table after retrieving the unread publications.
"""
def getUnreadPublications(self, delete_unread_entries=True):
conn = self.conn
cursor = conn.cursor()
cursor.execute(
'SELECT * FROM publications AS p LEFT JOIN unread AS u ON u.id=p.id;')
publications = cursor.fetchall()
if delete_unread_entries:
cursor.execute('DELETE FROM unread;')
conn.commit()
cursor.close()
publicationObjects = []
for p in publications:
publicationObjects.append(
Publication(p[0], p[1], p[2], p[3], p[4]))
return publicationObjects
However, in retrospect, it might make more sense to just have an additional column in the publications table as to whether its unread.
The analyze_publications function analyzes a publication to determine if it mentions any structural biology methods. It uses FAISS for embedding-based retrieval and the LLM for query evaluation.
def analyze_publication(self, text_embeddings: []):
"""poses a question about the document, processes the result and returns it
NOTE: for now, only uses the hackathon question, might add more later
Args:
text_embeddings ([]): list of (embedding, text) pairs from document to be analyzed
Returns:
bool: classification of response to query as positive (True) or negative (False)
str: response from chatGPT
"""
# NOTE: These very likely need to change
open_ai_emb = OpenAIEmbeddings()
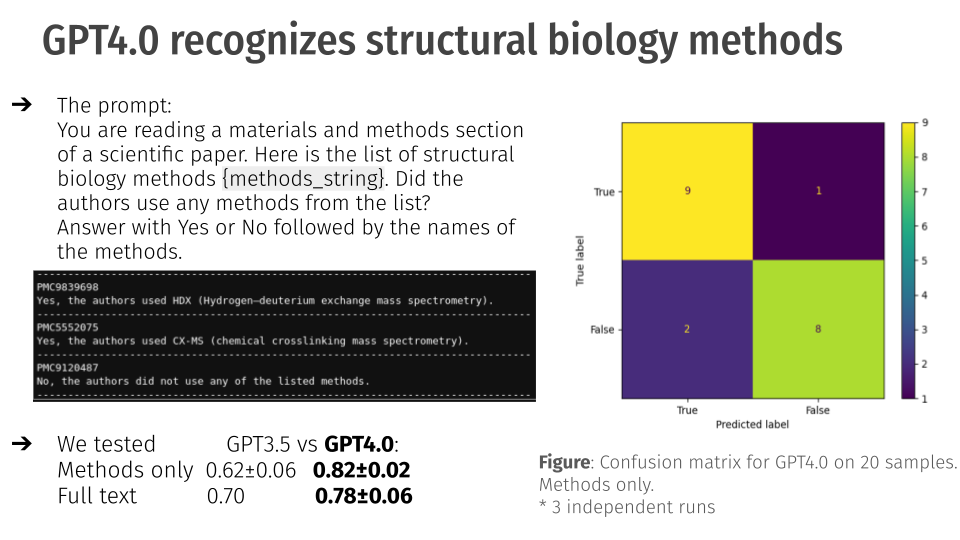
query = get_qbi_hackathon_prompt(METHODS_KEYWORDS)
faiss_index = FAISS.from_embeddings(text_embeddings=text_embeddings, embedding=open_ai_emb)
response = self.llm.evaluate_queries(faiss_index, query)[0]
return self.classify_response(response), response
We utilized several keywords to assist the LLM in identifing certain methodologies. We used the get_qbi_hackathon_prompt function to format our prompt using the keywords. They can be seen below as a guide:
# A list of abbreviated names and synonyms
# for various biophysical methonds
# that are typically used for integrative modeling
METHODS_KEYWORDS = {
'CX-MS': [
'cross-link', 'crosslink',
'XL-MS', 'CX-MS', 'CL-MS', 'XLMS', 'CXMS', 'CLMS',
"chemical crosslinking mass spectrometry",
'photo-crosslinking', 'crosslinking restraints',
'crosslinking-derived restraints', 'chemical crosslinking',
'in vivo crosslinking', 'crosslinking data',
],
'HDX': [
'Hydrogen–deuterium exchange mass spectrometry',
'Hydrogen/deuterium exchange mass spectrometry'
'HDX', 'HDXMS', 'HDX-MS',
],
'EPR': [
'electron paramagnetic resonance spectroscopy',
'EPR', 'DEER',
"Double electron electron resonance spectroscopy",
],
'FRET': [
'FRET',
"forster resonance energy transfer",
"fluorescence resonance energy transfer",
],
'AFM': [
'AFM', "atomic force microscopy",
],
'SAS': [
'SAS', 'SAXS', 'SANS', "Small angle solution scattering",
"solution scattering", "SEC-SAXS", "SEC-SAS", "SASBDB",
"Small angle X-ray scattering", "Small angle neutron scattering",
],
'3DGENOME': [
'HiC', 'Hi-C', "chromosome conformation capture",
],
'Y2H': [
'Y2H',
"yeast two-hybrid",
],
'DNA_FOOTPRINTING': [
"DNA Footprinting",
"hydroxyl radical footprinting",
],
'XRAY_TOMOGRAPHY': [
"soft x-ray tomography",
],
'FTIR': [
"FTIR", "Infrared spectroscopy",
"Fourier-transform infrared spectroscopy",
],
'FLUORESCENCE': [
"Fluorescence imaging",
"fluorescence microscopy", "TIRF",
],
'EVOLUTION': [
'coevolution', "evolutionary covariance",
],
'PREDICTED': [
"predicted contacts",
],
'INTEGRATIVE': [
"integrative structure", "hybrid structure",
"integrative modeling", "hybrid modeling",
],
'SHAPE': [
'Hydroxyl Acylation analyzed by Primer Extension',
]
}
def get_qbi_hackathon_prompt(keywords: dict) -> str:
"""
Returns a prompt that was initially developed
during the QBI Hackathon.
"""
if len(keywords) == 0:
raise(ValueError("Keywords dict can't be empty"))
methods_string = keywords_dict_to_string(keywords)
prompt = (
"You are reading a materials and methods section "
"of a scientific paper. "
f"Here is the list of methods {methods_string}.\n\n"
"Did the authors use any of them? "
"Answer Yes or No, followed by the name(s) of methods. "
"Use only abbreviations."
)
return prompt
We were also able to ask questions directly about the papers using the evaluate_queries method. We utilized the RetrievalQA capability in Langchain to provide retrieval augmentation for the response generation.
def evaluate_queries(self, embedding, queries):
chatbot = RetrievalQA.from_chain_type(
llm=self.llm,
chain_type="stuff",
retriever=embedding.as_retriever(search_type="similarity", search_kwargs={"k":3})
)
template = """ {query}? """
response = []
for q in queries:
prompt = PromptTemplate(
input_variables=["query"],
template=template,
)
response.append(chatbot.run(
prompt.format(query=q)
))
return response
Results
The results after completing the project were quite successful. Due to the nature of LLMs, we know that we need to test the accuracy to see if it is a useful tool. Using GPT 4.0, the developed system had an accuracy of .82 plus or minus 0.02 on a 95% confidence interval. Out of 20 runs, it got 17 correct, and provided two false positives and one false negative. This was more successful than GPT 3.5 which had a lower accuracy.
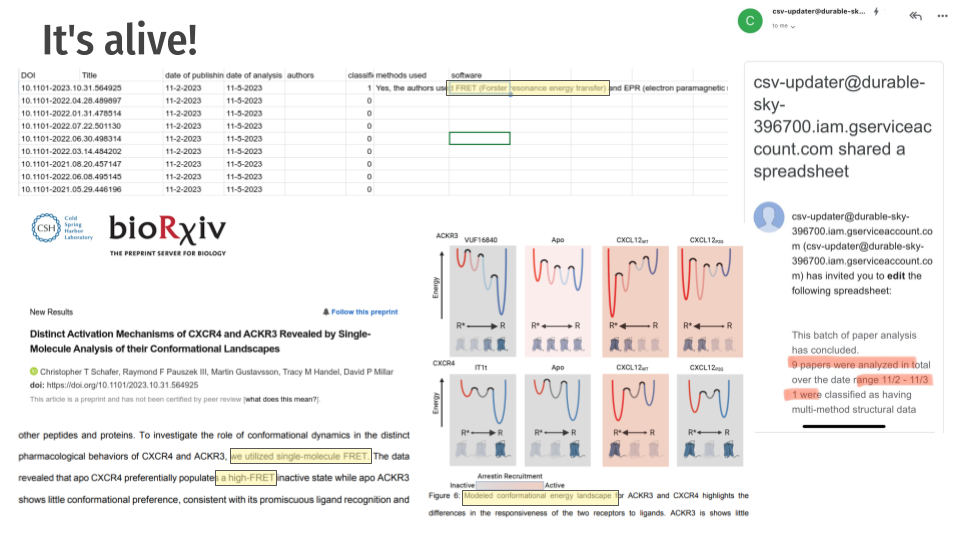
The system filled out a spreadsheet with information about the papers including the methods and technologies used and provided updates through email.
This project was a very fun hackathon project to work on and our team ended up winning first place!


Key Takeaways
There were several key takeaways from this event.
Structuring a Project Based on Requirements
One of the most critical aspects of this hackathon was learning how to structure a project effectively to meet specific requirements.
- Requirement Analysis: Understanding the core problem of data scarcity in the Protein Data Bank and how to address it with automation.
- Clear Objectives: Defining clear goals such as automating the data extraction and annotation process, ensuring metadata accuracy, and facilitating integration with existing databases.
- Modular Design: Breaking down the project into manageable components like the bioRxiv scraper, document analyzer, and database management. This modular approach ensured that each part could be developed, tested, and integrated independently.
Quick Delegation and Integration of Separate Work
Effective teamwork was another major takeaway.
- Role Assignment: Team members were assigned roles based on their expertise. For instance, some focused on backend development (like database management), others on the AI/ML components (like embeddings and LLM interaction), and others on integration and deployment.
- Regular Sync-Ups: The team held frequent check-ins to ensure everyone was on the same page, discuss progress, and troubleshoot any issues collaboratively.
- Integration Strategy: A clear integration plan was established early on. This involved setting up version control (e.g., using Git) and defining API contracts between different modules to ensure smooth integration.
Working with LLM Technologies
The hackathon provided hands-on experience with various LLM technologies and tools.
- Vector Embeddings: Understanding how to convert text into vector embeddings using OpenAI’s embeddings and storing them in a vector database like Lantern Vector+postgres. This step was crucial for efficient retrieval and querying.
- FAISS Context Retrieval: Learning how to use FAISS for context retrieval based on cosine similarity. This involved setting up FAISS and using it to fetch relevant document fragments.
- Langchain for LLM Utilities: Utilizing Langchain to handle various LLM utilities, making the interaction with the GPT-4.0 API more seamless and efficient.
- Automating Data Annotation: Developing scripts that leverage GPT-4.0 to automate the process of annotating scientific papers, extracting relevant metadata, and updating databases.
The repository with all of the code can be found below:
Enjoy Reading This Article?
Here are some more articles you might like to read next: